Third-party packages¶
This page lists the third-party packages/modules built upon the MyVariant.info services.
MyVariant python module¶
“myvariant” is an easy-to-use Python wrapper to access MyVariant.info services.
You can install it easily using either pip or easy_install:
pip install myvariant #this is preferred
or:
easy_install myvariant
This is a brief example:
In [1]: import myvariant
In [2]: mv = myvariant.MyVariantInfo()
In [3]: mv.getvariant('chr1:g.35367G>A')
Out[3]:
{'_id': 'chr1:g.35367G>A',
'_version': 1,
'cadd': {'alt': 'A',
'annotype': 'NonCodingTranscript',
'bstatistic': 994,
'chmm': {'bivflnk': 0.0,
'enh': 0.0,
'enhbiv': 0.0,
'het': 0.0,
'quies': 1.0,
'reprpc': 0.0,
'reprpcwk': 0.0,
'tssa': 0.0,
'tssaflnk': 0.0,
'tssbiv': 0.0,
'tx': 0.0,
'txflnk': 0.0,
'txwk': 0.0,
'znfrpts': 0.0},
'chrom': 1,
'consdetail': 'non_coding_exon,nc',
'consequence': 'NONCODING_CHANGE',
'consscore': 5,
'cpg': 0.03,
'dna': {'helt': -2.04, 'mgw': 0.01, 'prot': 1.54, 'roll': -0.63},
'encode': {'exp': 31.46, 'h3k27ac': 23.44, 'h3k4me1': 23.8, 'h3k4me3': 8.6},
'exon': '2/3',
'fitcons': 0.577621,
'gc': 0.48,
'gene': {'cds': {'cdna_pos': 476, 'rel_cdna_pos': 0.4},
'feature_id': 'ENST00000417324',
'gene_id': 'ENSG00000237613',
'genename': 'FAM138A'},
'gerp': {'n': 1.29, 's': -0.558},
'isknownvariant': 'FALSE',
'istv': 'FALSE',
'length': 0,
'mapability': {'20bp': 0, '35bp': 0},
'min_dist_tse': 122,
'min_dist_tss': 707,
'mutindex': 70,
'phast_cons': {'mammalian': 0.003, 'primate': 0.013, 'vertebrate': 0.003},
'phred': 1.493,
'phylop': {'mammalian': -0.155, 'primate': 0.151, 'vertebrate': -0.128},
'pos': 35367,
'rawscore': -0.160079,
'ref': 'G',
'scoresegdup': 0.99,
'segway': 'D',
'type': 'SNV'},
'snpeff': {'ann': [{'effect': 'non_coding_exon_variant',
'feature_id': 'NR_026818.1',
'feature_type': 'transcript',
'gene_id': 'FAM138A',
'gene_name': 'FAM138A',
'hgvs_c': 'n.476C>T',
'putative_impact': 'MODIFIER',
'rank': '2',
'total': '3',
'transcript_biotype': 'Noncoding'},
{'effect': 'non_coding_exon_variant',
'feature_id': 'NR_026820.1.2',
'feature_type': 'transcript',
'gene_id': 'FAM138F.2',
'gene_name': 'FAM138F',
'hgvs_c': 'n.476C>T',
'putative_impact': 'MODIFIER',
'rank': '2',
'total': '3',
'transcript_biotype': 'Noncoding'}]},
'vcf': {'alt': 'A', 'position': '35367', 'ref': 'G'}}
See https://pypi.python.org/pypi/myvariant for more details.
MyVariant R package¶
An R wrapper for the MyVariant.info API is available in Bioconductor since v3.2. To install:
source("https://bioconductor.org/biocLite.R")
biocLite("myvariant")
To view documentation for your installation, enter R and type:
browseVignettes("myvariant")
For more information, visit the Bioconductor myvariant page.
MyVariant Node.js package¶
myvariantjs is a Node.js wrapper for the MyVariant.info API, developed and maintained by Larry Hengl. To install:
npm install myvariantjs --save
Some brief usage examples:
var mv = require('myvariantjs');
mv.getvariant('chr9:g.107620835G>A');
mv.getvariant('chr9:g.107620835G>A', ['dbnsfp.genename', 'cadd.phred']);
mv.getvariants("chr1:g.866422C>T,chr1:g.876664G>A,chr1:g.69635G>C"); // string of delimited ids
mv.getvariants(["chr1:g.866422C>T", "chr1:g.876664G>A","chr1:g.69635G>C"]);
mv.query("chr1:69000-70000", {fields:'dbnsfp.genename'});
mv.query("dbsnp.rsid:rs58991260", {fields:'dbnsfp'});
mv.querymany(['rs58991260', 'rs2500'], 'dbsnp.rsid');
mv.querymany(['RCV000083620', 'RCV000083611', 'RCV000083584'], 'clinvar.rcv_accession');
For more information, visit its API and usage docs, and its github code repository.
You can also check out this neat demo application developed by Larry using this myvariantjs package.
Another MyVariant.info python module¶
This is another python wrapper of MyVariant.info services created by Brian Schrader. The repository is available here.
You can install this package with PyPI like this:
pip install myvariant-api
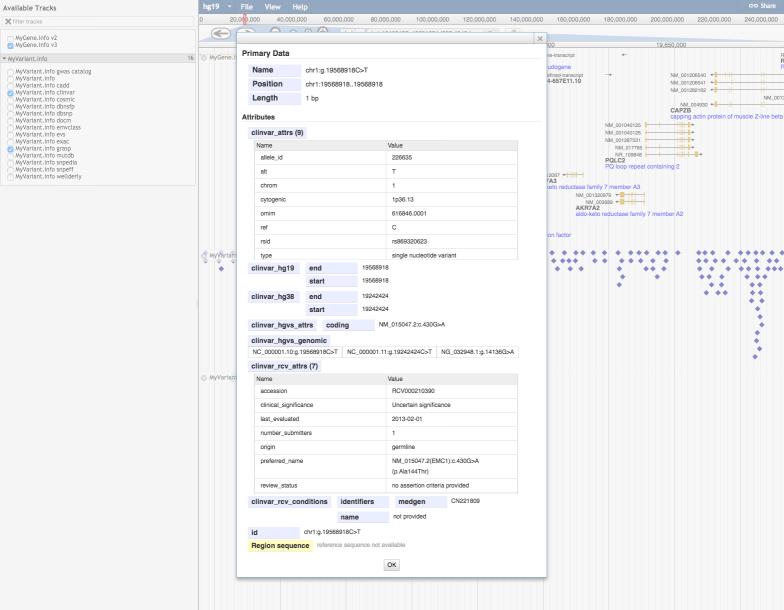
A JBrowse plugin for MyVariant.info and MyGene.info¶
JBrowse provides a fast, embeddable genome browser built completely with JavaScript and HTML5.
Colin from the JBrowse team made a very nice plugin to visualize the gene and variant annotations in JBrowse Genome Browser, using the data served from both MyGene.info and MyVariant.info APIs.
Live demo
To see it live, here is the demo site. It has been tested with hg38, hg19, and zebrafish and has mygene.info and myvariant.info integrations
Source code
A screenshot